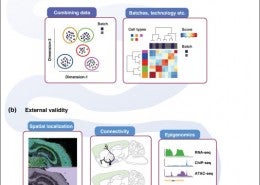
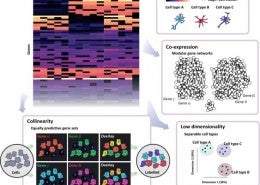
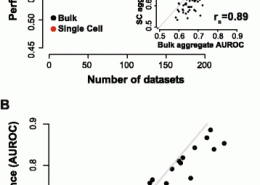
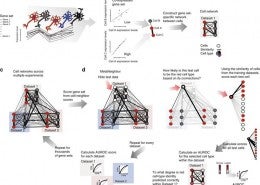
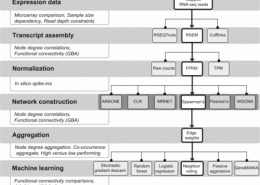
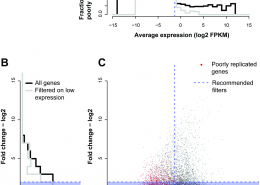
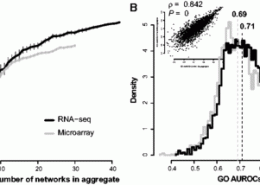
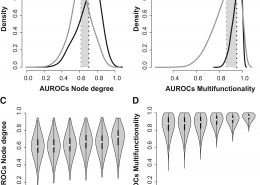
Our work centers on characterizing gene networks to understand cell identity and disease. We particularly focus on gene networks derived from expression data through the combination of hundreds or thousands of experiments. Some of the issues surrounding network analysis are discussed in the opinion pieces:
https://f1000research.com/articles/1-14/v1
https://www.cell.com/trends/genetics/fulltext/S0168-9525(18)30128-8
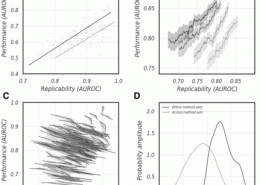
Single-cell RNA-seq (scRNA-seq) has revolutionized the way we look at the transcriptome. Our work has focused on characterizing cellular identity and assessing replicability of signals.
Crow, M., Gillis, J. (June 2019) Single cell RNA-sequencing: replicability of cell types. Current Opinion in Neurobiology, 56. pp. 69-77. ISSN 09594388 (ISSN)
Crow, M., Gillis, J. (August 2018) Co-expression in Single-Cell Analysis: Saving Grace or Original Sin? Trends Genet. ISSN 0168-9525 (Print)0168-9525
Kalish, B. T., Cheadle, L., Hrvatin, S., Nagy, M. A., Rivera, S., Crow, M., Gillis, J., Kirchner, R., Greenberg, M. E. (January 2018) Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc Natl Acad Sci U S A, 115 (5). E1051-E1060. ISSN 0027-8424
Crow, M., Paul, A., Ballouz, S., Huang, Z. J., Gillis, J. (May 2016) Exploiting single-cell expression to characterize co-expression replicability. Genome Biol, 17 (1). p. 101. ISSN 1474-760X (Electronic)1474-7596 (Linking) (Public Dataset)
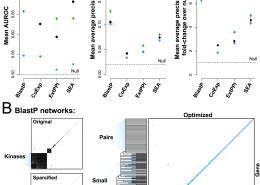
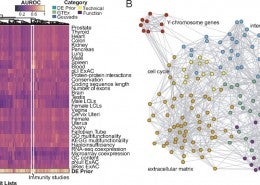
Gene co-expression network analysis is a field merging network theory and transcriptomics. Co-expression is meant to reflect co-regulation, co-functionality and co-variation. We have shown the utility of co-expression, in particular meta-analytic co-expression, in a variety of applications.
Crow, M., Paul, A., Ballouz, S., Huang, Z. J., Gillis, J. (May 2016) Exploiting single-cell expression to characterize co-expression replicability. Genome Biol, 17 (1). p. 101. ISSN 1474-760X (Electronic)1474-7596 (Linking) (Public Dataset)
Ballouz, S., Gillis, J. (April 2016) AuPairWise: A Method to Estimate RNA-Seq Replicability through Co-expression. PLoS Comput Biol, 12 (4). e1004868. ISSN 1553-7358 (Electronic)1553-734X (Linking)
Ballouz, S., Verleyen, W., Gillis, J. (February 2015) Guidance for RNA-seq co-expression network construction and analysis: safety in numbers. Bioinformatics. ISSN 1367-4803
The ‘guilt by association’ principle underlies many gene function prediction algorithms. Our research focuses on identifying limitations in the GBA approach and making fundamental improvements to its operation for the interpretation of neuropsychiatric genomics data.
Ballouz, S., Weber, M., Pavlidis, P., Gillis, J. (February 2017) EGAD: ultra-fast functional analysis of gene networks. Bioinformatics, 33 (4). pp. 612-614. ISSN 1367-4803
Verleyen, W., Ballouz, S., Gillis, J. (March 2015) Measuring the wisdom of the crowds in network-based gene function inference.Bioinformatics, 31 (5). pp. 745-752. ISSN 1367-4803
Gillis, J., Pavlidis, P. (March 2012) “Guilt by Association” Is the Exception Rather Than the Rule in Gene Networks. PLoS Computational Biology, 8 (3). ISSN 1553-734X
Gillis, J., Pavlidis, P. (2011) The role of indirect connections in gene networks in predicting function. Bioinformatics, 27 (13). pp. 1860-1866. ISSN 13674803 (ISSN)
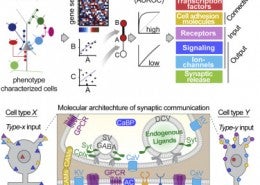
To enhance our understanding of cognitive disorders and the brain more generally, we have integrated genomics and neuroscience research.
Crow, M., Paul, A., Ballouz, S., Huang, Z. J., Gillis, J. (February 2018) Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat Commun, 9 (1). p. 884. ISSN 2041-1723
Kalish, B. T., Cheadle, L., Hrvatin, S., Nagy, M. A., Rivera, S., Crow, M., Gillis, J., Kirchner, R., Greenberg, M. E. (January 2018) Single-cell transcriptomics of the developing lateral geniculate nucleus reveals insights into circuit assembly and refinement. Proc Natl Acad Sci U S A, 115 (5). E1051-E1060. ISSN 0027-8424
Paul, A., Crow, M., Raudales, R., He, M., Gillis, J., Huang, Z. J. (2017) Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell.
Ballouz, S., Gillis, J. (July 2017) Strength of functional signature correlates with effect size in autism. Genome Med, 9 (1). p. 64. ISSN 1756-994x
McCarthy, S. E., Gillis, J., Kramer, M., Lihm, J., Yoon, S., Berstein, Y., Mistry, M., Pavlidis, P., Solomon, R., Ghiban, E., Antoniou, E., Kelleher, E., O’Brien, C., Donohoe, G., Gill, M., Morris, D. W., McCombie, W. R., Corvin, A. (June 2014) De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Molecular Psychiatry, 19 (6). pp. 652-658. ISSN 14765578
Gene multifunctionality is a pervasive bias in functional genomics. Our work has focused on evaluating how this bias impacts the generation of biologically non-specific results as well as highly fragile significances in a variety of fields.
Crow, M., Lim, N., Ballouz, S., Pavlidis, P., Gillis, J. (March 2019) Predictability of human differential gene expression. Proc Natl Acad Sci U S A. ISSN 0027-8424
Ballouz, S., Dobin, A., Gingeras, T. R., Gillis, J. (May 2018) The fractured landscape of RNA-seq alignment: the default in our STARs.Nucleic Acids Res. ISSN 0305-1048
Ballouz, S., Pavlidis, P., Gillis, J. (October 2016) Using predictive specificity to determine when gene set analysis is biologically meaningful. Nucleic Acids Res. ISSN 1362-4962 (Electronic)0305-1048 (Linking)
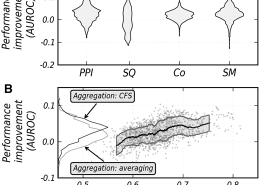
Verleyen, W., Ballouz, S., Gillis, J. (April 2016) Positive and negative forms of replicability in gene network analysis. Bioinformatics, 32 (7). pp. 1065-73. ISSN 1367-4811 (Electronic)1367-4803 (Linking)
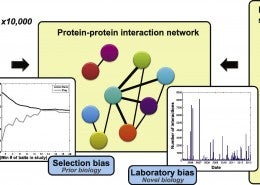
Gillis, J., Ballouz, S., Pavlidis, P. (January 2014) Bias tradeoffs in the creation and analysis of protein-protein interaction networks.Journal of Proteomics, 100. pp. 44-54.
Gillis, J., Pavlidis, P. (February 2013) Assessing identity, redundancy and confounds in Gene Ontology annotations over time.Bioinformatics, 29 (4). pp. 476-82. ISSN 1367-4811 (Electronic)1367-4803 (Linking)
Gillis, J., Pavlidis, P. (2013) Characterizing the state of the art in the computational assignment of gene function: lessons from the first critical assessment of functional annotation (CAFA). BMC Bioinformatics, 14 (Suppl). S15. ISSN 1471-2105 (Electronic)1471-2105 (Linking)